Abstract
The gold standard for the diagnosis of MDS relies on morphologic alterations hampered by a great deal of subjectivity. Cytogenetic and clinical features allow for clinical classifications predictive of survival. However, with a few exceptions (SF3B1MT, del5q, and certain balanced translocations), neither classic histo-morphology nor prognostic scoring systems (e.g., IPSS-R) are reflective of pathogenic underpinnings. To date supervised analyses of mutational data did not succeed to produce profiles specific or predictive of traditional disease sub-entities. Large cohorts with clinical annotation and a sufficient follow-up allow for innovative biostatistical approaches to subgroup patients according to molecular profiles. Objective operator-independent subcategorization may be congruent with common pathogenic links, rational applications of targeted therapeutics and better prognostications.
We hypothesized that machine-learning (ML) strategies used for analysis of mutational/cytogenetic profiles will enable recognition of invariant disease subcategories according to their molecular configurations. Herein, we compiled a meta-analytic database (our cohorts and publicly available sources) of 3,011 MDS (median age 71yrs) and 6,788 pAML/sAML. Results of deep targeted sequencing of a panel of 55 myeloid mutations were collected together with cytogenetics. We then performed unsupervised analysis of MDS and AML patients using Bayesian Latent Class Analysis (BLCA). A consensus matrix was then clustered using Ward's criteria to generate final cluster assignment based on the highest silhouette value. To identify genomic signatures, we used Random Forest classification and extracted mutations with highest global importance indicated by mean decrease in accuracy.
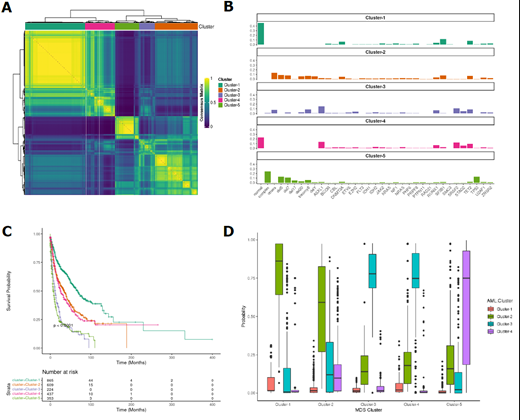
Using BLCA we identified 5 unique genomic clusters (GCs) with 3 distinct prognostic outcomes [low risk (LR), intermediate risk (Int), and high risk (HR)] that were validated by survival analysis (Fig.1A,B). The LR group included GC-1 and was characterized by the highest prevalence of normal cytogenetics (100%) and SF3B1 MT (25%) with co-occurring DNMT3A MT (14%), and absence of ASXL1 MT, ETV6 MT, STAG2 MT, TP53 MT, and complex/abnormal cytogenetics. Int group included GC-2 and GC-4. GC-2 was characterized by a higher percentage of abnormal cytogenetics cases than LR group and absence of STAG2 MT, SRSF2 MT, ASXL1 MT, TP53 MT, and normal/complex cytogenetics. GC-4 had the highest frequency of SRSF2 MT (52%) with co-occurring ASXL1 MT (59%), TET2 MT (40%), normal karyotype, and absence of complex/abnormal cytogenetics. Finally, HR included GC-3 and GC-5. GC-3 included ASXL1 MT (67%) with co-occurring SRSF2 MT (47%), TET2 MT (37%), STAG2 MT (22%), and absence of normal cytogenetics. GC- 5 had the highest frequency of -5/del(5q) (50%), -7/del(7q) (43%), -17/del(17p) (16%) and the highest odds of complex karyotype (92%) as well as TP53 MT (48%). Paralleling the genomic ML-based clustering, the clinical relevance of these subgroups was reflected in significantly different survivals [median (95% CI)]: i) GC-1 [69 (59-80)], ii) GC-2 [35 (29-41)], iii) GC-3 [12 (10-16)], GC-4 [27 (22-34)], and GC-5 [9 (7-11)] months (Fig.1C). We then classified the MDS cohort according to the recently published and validated AML GCs (Awada et al Blood 2021) to investigate overlapping genomic features. Overall, 90% of MDS GC-1 and 67% of MDS GC-2 had the same molecular architecture of AML GC-2 and 70% of MDS GC-5 had the same molecular features of AML GC-4. In addition, 98% of MDS GC-3 and 92% of MDS GC-4 had the same features of AML GC-3 (Fig.1D).
In sum, we propose a novel objective molecular classification of MDS and related diseases that allows subgrouping of patients according to shared pathogenesis for a better prognostic resolution without errors derived from subjectivity. The model was then internally and externally validated using a cohort of 200 cases. Results of a validation cohort and online URL site of molecular clustering will be presented at the meeting.
Balasubramanian: Servier Pharmaceuticals: Research Funding. Patel: Alexion: Consultancy, Other: educational talks, Speakers Bureau; Apellis: Consultancy, Other: educational talks, Speakers Bureau. Carraway: Celgene, a Bristol Myers Squibb company: Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding, Speakers Bureau; Astex: Other: Independent review committee; Novartis: Honoraria, Membership on an entity's Board of Directors or advisory committees, Speakers Bureau; Agios: Honoraria, Membership on an entity's Board of Directors or advisory committees, Speakers Bureau; Takeda: Other: Independent review committee; Bristol Myers Squibb: Honoraria, Membership on an entity's Board of Directors or advisory committees, Speakers Bureau; Jazz: Honoraria, Membership on an entity's Board of Directors or advisory committees, Speakers Bureau; AbbVie: Other: Independent review committee; Stemline: Honoraria, Membership on an entity's Board of Directors or advisory committees, Speakers Bureau. Haferlach: MLL Munich Leukemia Laboratory: Other: Part ownership. Maciejewski: Regeneron: Consultancy; Novartis: Consultancy; Alexion: Consultancy; Bristol Myers Squibb/Celgene: Consultancy.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal